Resources and Tools
Our software, tools, datasets, etc. are open-source, and free for anyone around the world to use and modify. We strive to make resources that are high quality in every aspect: cleanly written, robustly constructed and tested, well-documented, easy-to-use, accessible, customizable, and as effective as possible in real-world use.
Data portal
 Human Cell Atlas
Human Cell AtlasSingle cell atlas of different parts of the human eye generated by combining previously published and newly generated datasets.
 Mouse Cell Atlas
Mouse Cell AtlasA unified single-cell atlas of the mouse retina using the most comprehensive approximately 323,000 single cells from our in-house experiments complementing public datasets.
 Mouse Retina Single Cell Transcript Isoform Catalog
Mouse Retina Single Cell Transcript Isoform CatalogBy profiling the transcriptome of approximately 30,000 mouse retina cells with 1.54 billion Illumina short reads and 1.40 billion Oxford Nanopore Technologies long reads, we identify 44,325 transcript isoforms, with a notable 38% previously uncharacterized and 17% expressed exclusively in distinct cellular subclasses.
 Macaque Capture and Exome Database
Macaque Capture and Exome DatabaseThe macaque Capture and Exome database (mCED) provides access to genotype data of rhesus macaques collected from eight non-primate research centers.
Software and Tools
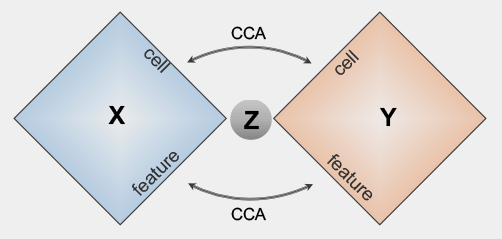 bindSCBi-order Integration of multi-omics Data from Single Cell sequencing technologies
bindSCBi-order Integration of multi-omics Data from Single Cell sequencing technologiesbindSC is an R package for single-cell multi-omic integration analysis. It is developed to address the challenge of single-cell multi-omic data integration that consists of unpaired cells measured with unmatched features across modalities.
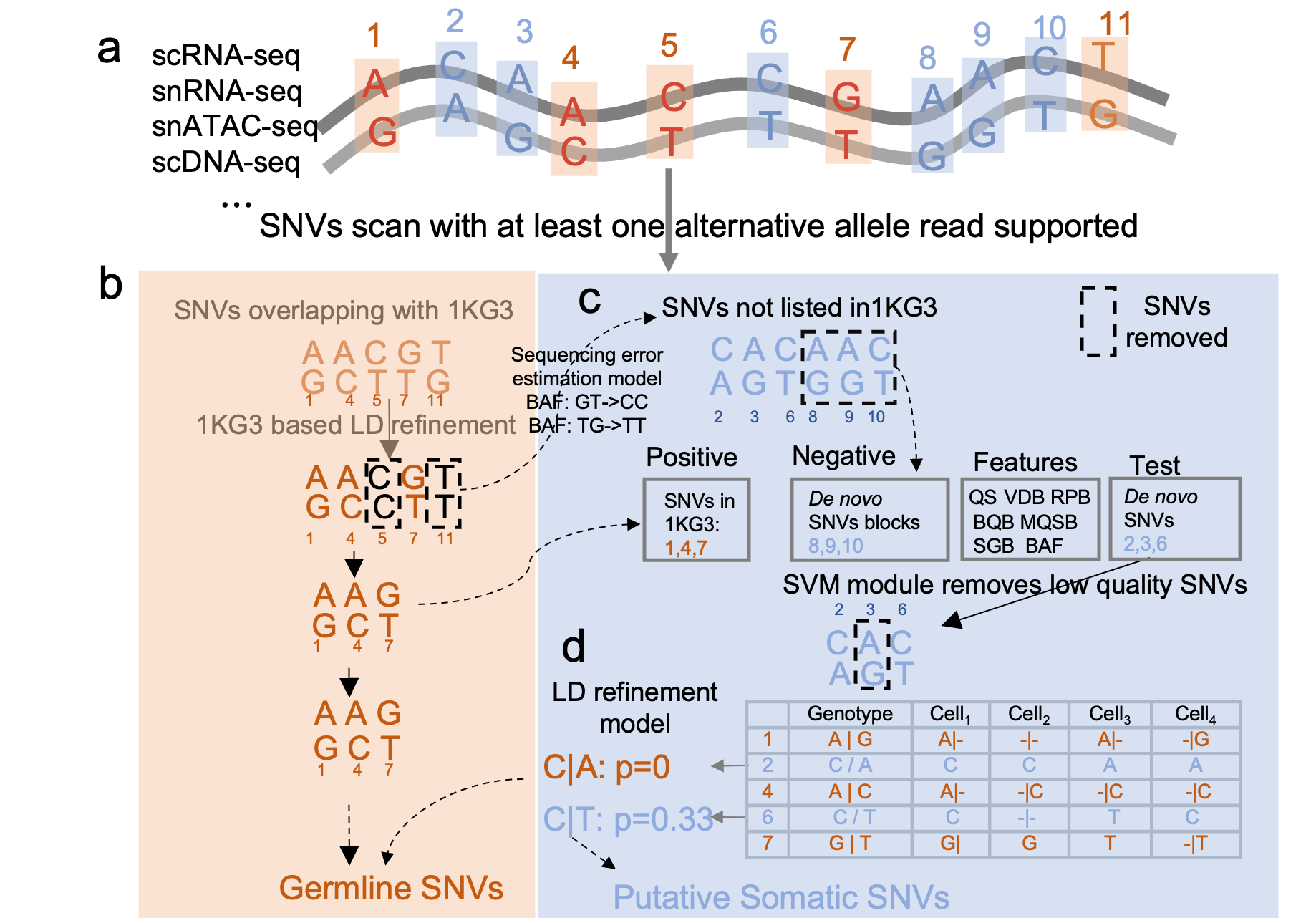 MonopogenSNV calling from single-cell sequencing data
MonopogenSNV calling from single-cell sequencing dataMonopogen is an analysis package for SNV calling from single-cell sequencing. Monopogen works on sequencing datasets generated from single cell RNA 10x 5’, 10x 3’, single ATAC-seq technoloiges, scDNA-seq etc.
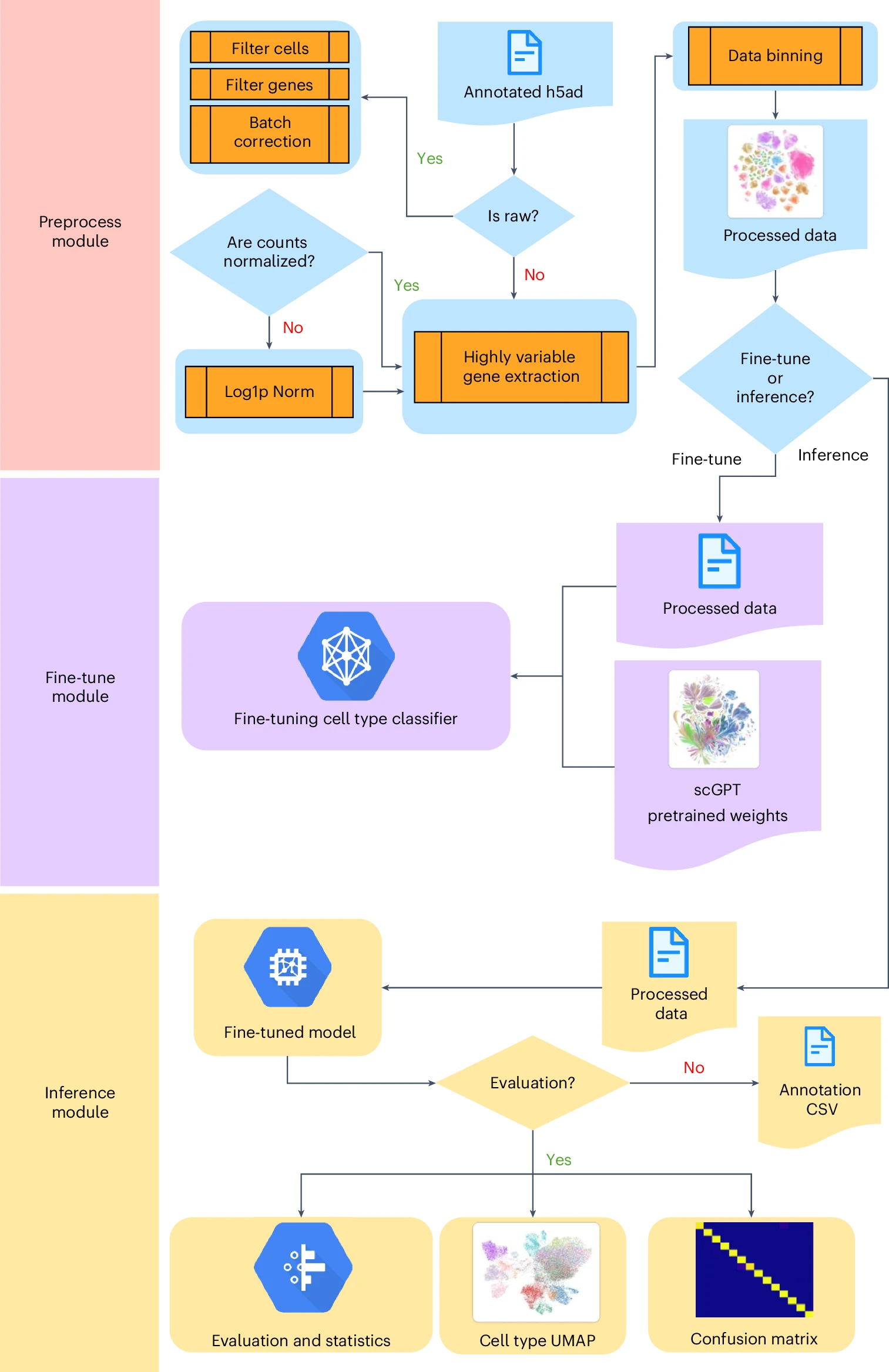 Fine-tune Protocol for eye-scGPTProtocol for doing fine-tuning on any single-cell annotations with scGPT
Fine-tune Protocol for eye-scGPTProtocol for doing fine-tuning on any single-cell annotations with scGPTThis protocol enables researchers to efficiently deploy scGPT for their own datasets. The provided tools, including a command-line script and Jupyter Notebook, simplify the customization and exploration of the model.
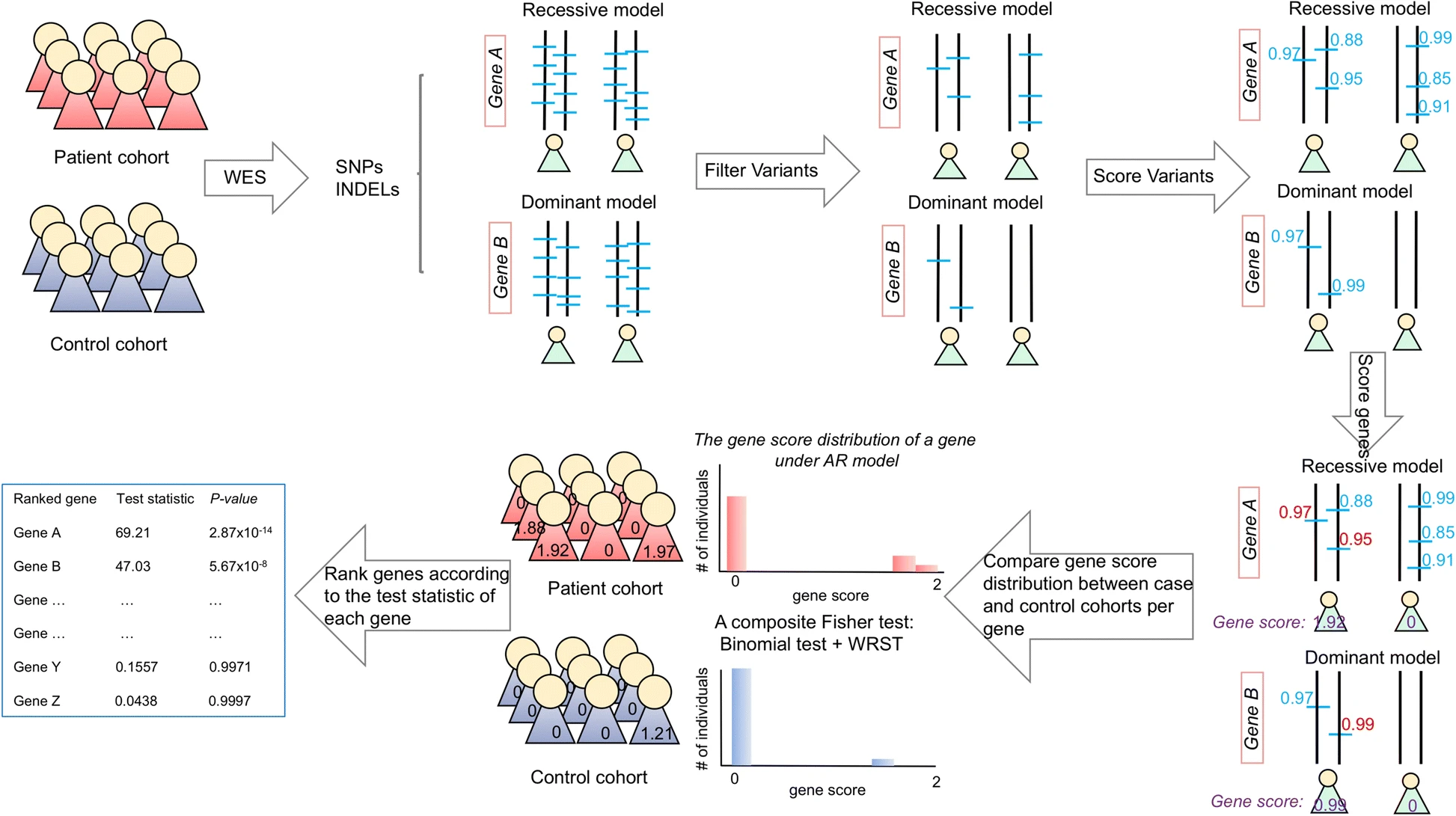 GRIPTA novel case-control analysis method for Mendelian disease gene discovery
GRIPTA novel case-control analysis method for Mendelian disease gene discoveryA novel method, the Gene Ranking, Identification and Prediction Tool (GRIPT), for performing case-control analysis of NGS data which is well-powered for disease gene discovery, especially for diseases with high locus heterogeneity.
Quick Links
Gulf Coast Cluster for Single Cell Omics
The GCC single cell omics cluster aims at leveraging the expertise at different institutions and maximize the interdisciplinary synergy of the group.
The eye biological network
The eye biological network will establish a comprehensive molecular and spatial atlas of the cells in the visual system across age and ethnicity groups. This resource will serve the foundation for further dissection of the function, interaction, and involvement of subtypes of cells in the visual function and diseases.
Network Coordinator: Rui Chen, Ph.D. Email
Single Cell Genomics Core
The Single Cell Genomics Core provides comprehensive services for RNA and chromatin profiling on single or a small number of cells.
The core is equipped with the 10x Genomics Chromium Platform, the Takara icell8 platform, and the Fluidigm C1 auto cell prep platforms. The combination of these platforms allows cost-effective and high throughput genomic profiling that matches with the experiments.
Academic Director: Rui Chen, Ph.D. Email
X-linked Inherited Retinal Disease Variant Curation Expert Panel
We will have established variant curation rules that can be applied for all X- linked IRD genes, completed the curation of these IRD genes, and established a framework that can be modified and applied to other genes affecting inherited retinal disease.
Ancestry Network for the Human Cell Atlas of the Eye
To understand the contribution of genetic background to the human visual system, this group will generate data from diverse ancestral groups, as part of the Human Cell Atlas project.
Internal Links
Genetics
References: UCSC Genome Browser
gnomAD browser
Ensembl
GeneCards
OMIM
DECIPHER
ClinGen
HGNC
NCBI
ClinVar
LOVD
Effect Predictor & others: REVEL
CADD
EVE
dbNSFP
spliceAI
VEP
AnnotSV
Position Converter
Single Cell
Tools: scRNA-Tools Seurat Cellxgene DoubletFinder DropletUtils SeuratDisk SoupX ScPred Scanpy dropkick CellRank scvi-tools ArchR
